This is from the Illumina #AGBT25 workshop about their ST tech.
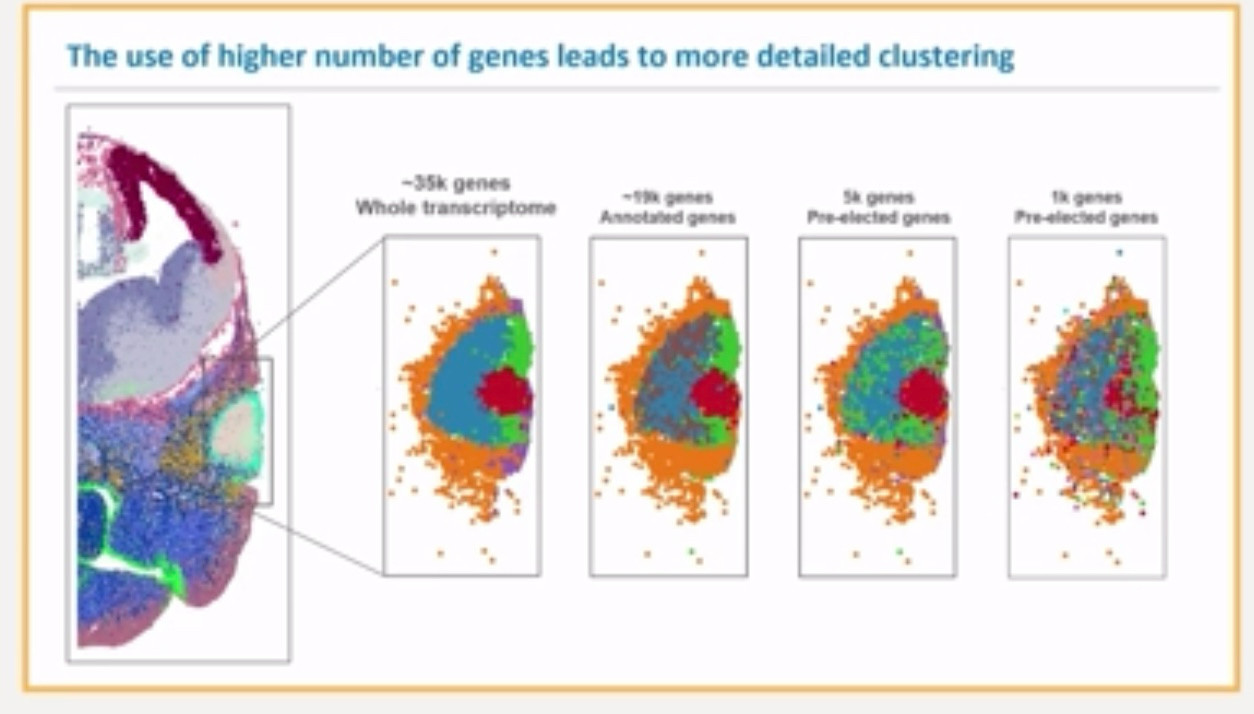
But, from our observations, under heavy lateral diffusion (real or simulated), clustering on large sets of genes (i.e. “whole transcriptome”) leads to niche- rather than cell-type clustering.
This might be problematic.
But, from our observations, under heavy lateral diffusion (real or simulated), clustering on large sets of genes (i.e. “whole transcriptome”) leads to niche- rather than cell-type clustering.
This might be problematic.
Comments
Very sparsely 2D-cultured cells (on chip) and measuring some very well known markers, would give a good baseline.
Note: lateral diffusion might happen during in-situ capture, but more important is the “diffuse background” that appears during prior steps. This actually drives most clustering artifacts.
I don’t share them bc I think the results were not super satisfactory. It is a completely open problem.