Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics?
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
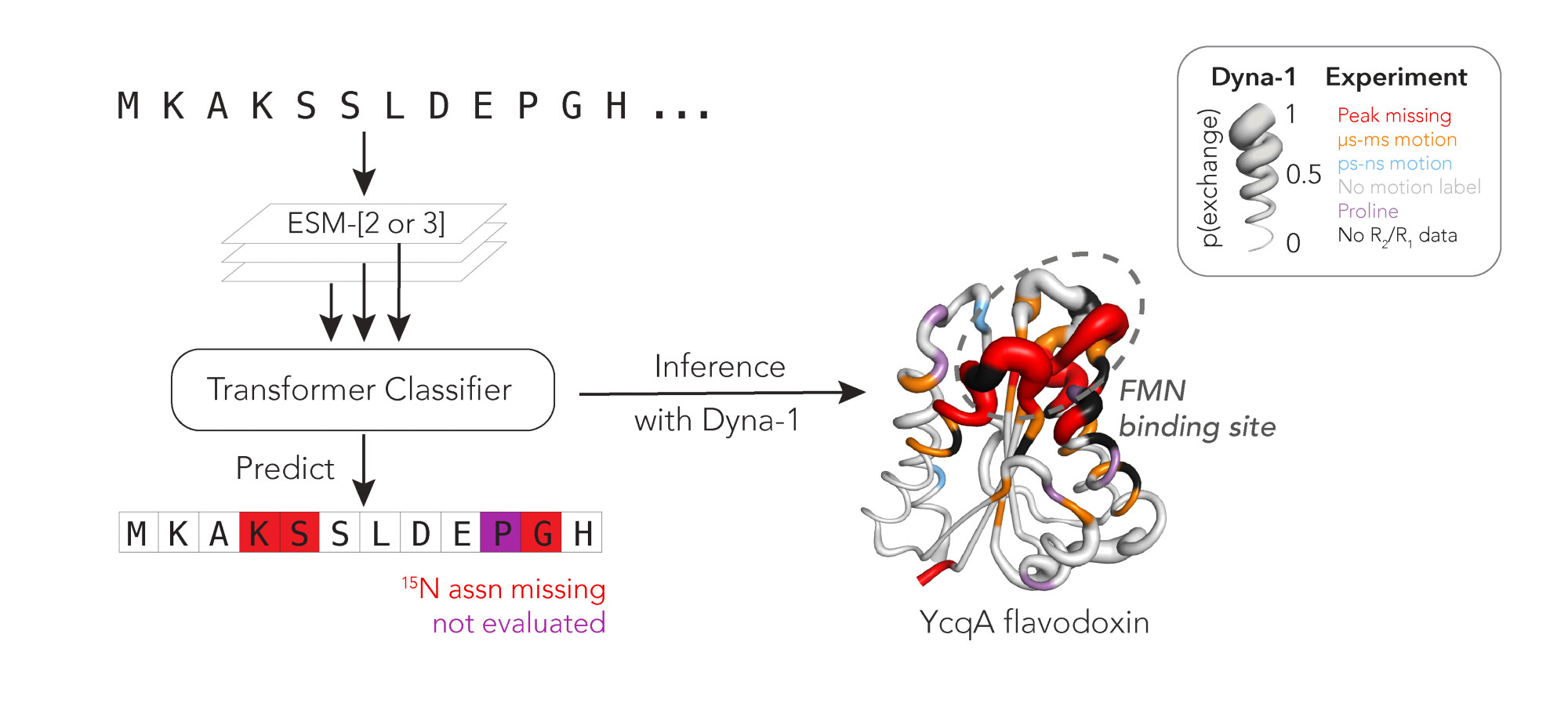
Comments
To me, the kicker things we want to predict are coordinated motions that have low prob. Saw hints of that with CypA. I don’t think people would think of that as flexibility …
That's Rex, a measurable from NMR. Rex increases when an atom moves at the µs-ms timescale.
There is no DB for that, so Hannah curated "RelaxDB"
Where do we find these chemical shift datasets? The BMRB!
Hannah curated another labeled dataset: the "mBMRB"
To start, 45% of the proteins had ≥1 NMR structure but no X-ray/EM structures
...and there is no difference in B-factors between assigned & missing aa's