In a new preprint led by @TheNikhilMilind, we explored a fascinating paradox:
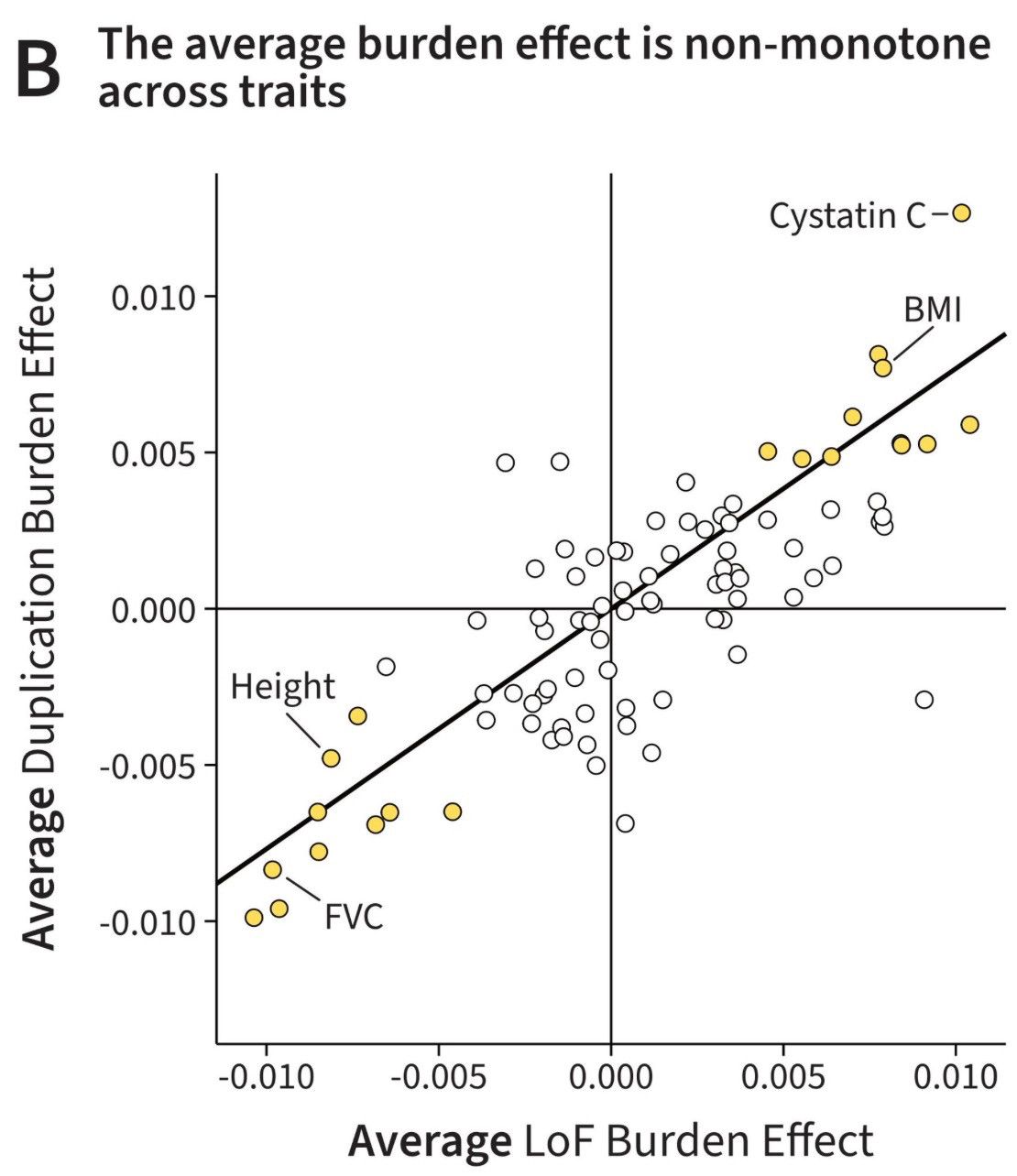
For many traits the number of duplications or loss-of-function (LoF) mutations is correlated with phenotype. Curiously, for most traits, the AVERAGE direction of LoFs and Dups is the SAME. Why?
For many traits the number of duplications or loss-of-function (LoF) mutations is correlated with phenotype. Curiously, for most traits, the AVERAGE direction of LoFs and Dups is the SAME. Why?
Comments
In a new preprint we explore why this happens:
@nikhilmilind.dev @courtsmithrun.bsky.social Julie Zhu @jeffspence.bsky.social
https://www.medrxiv.org/content/10.1101/2024.11.11.24317065v1
Take fitness for example - genes will be selected to be near optimum dosage, so deviation in either direction lowers fitness.
But somehow, aggregating this effect across a bunch of genes, results in a SAME-direction effect on average, genome-wide. Why?
Some top hits:
cc: @nikhilmilind.dev