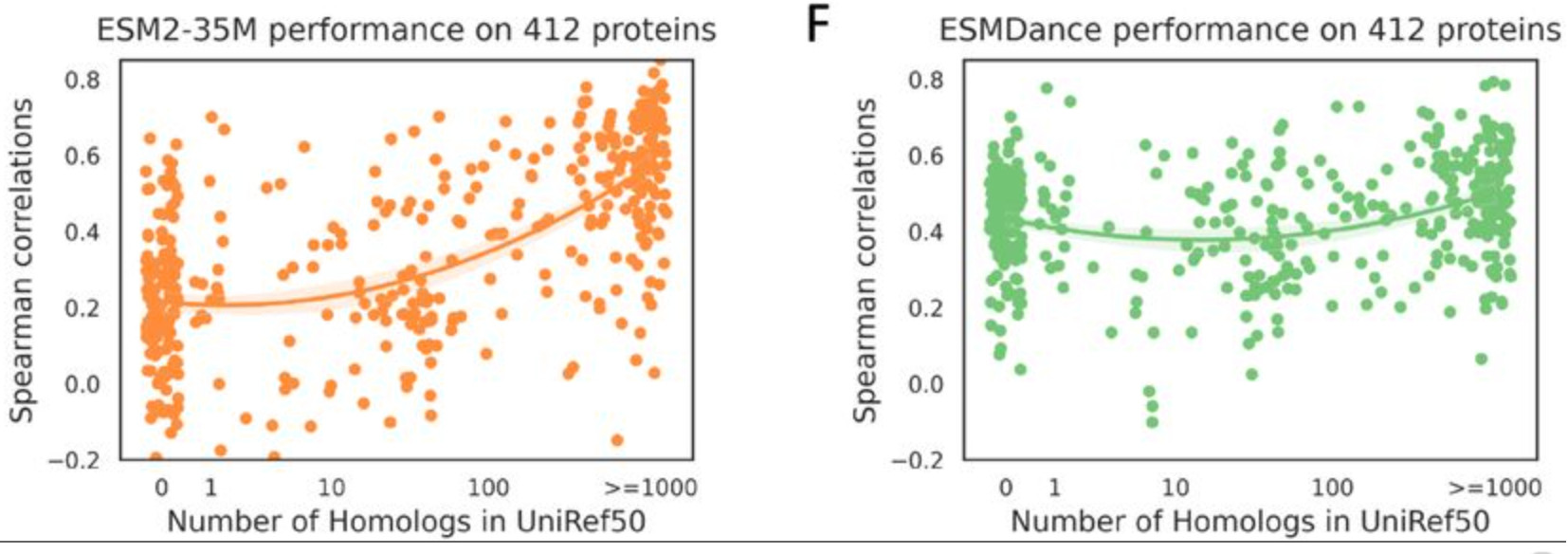
PLMs fine-tuned on protein structural dynamics data show improved zero-shot variant effect prediction. However, this improvement is lost as the number of homologs in the training set increases. The data for fine-tuning includes MD and NMA data
https://www.biorxiv.org/content/10.1101/2024.10.11.617911v2
https://www.biorxiv.org/content/10.1101/2024.10.11.617911v2
Comments