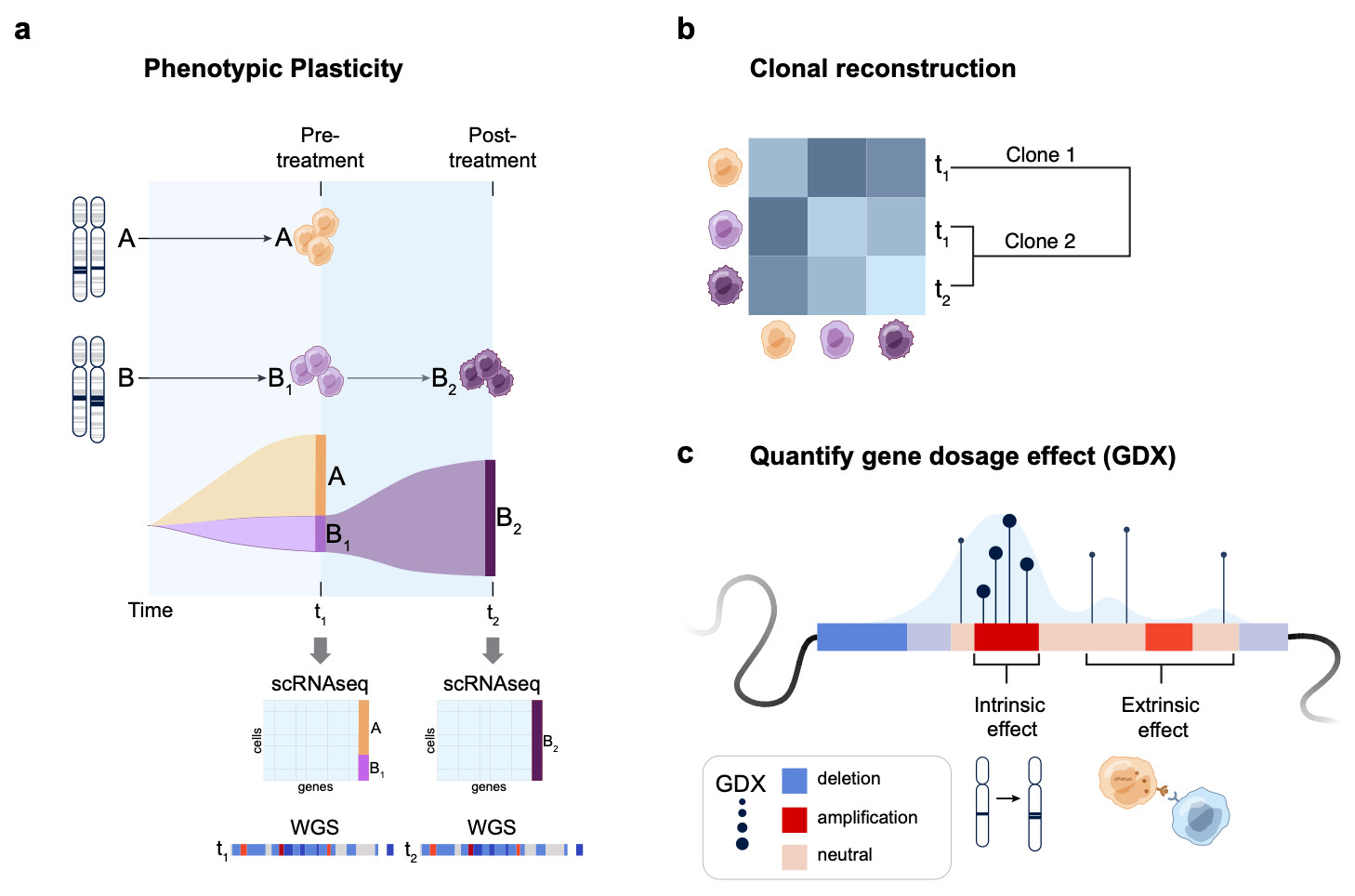
How it works: #Echidna models single-cell RNA & bulk WGS data using a Bayesian hierarchical framework. Key features:
Deconvolves CNA profiles
Tracks clones & their phenotypic states over time
Introduces GDX: a metric quantifying gene dosage effects! 3/
Deconvolves CNA profiles
Tracks clones & their phenotypic states over time
Introduces GDX: a metric quantifying gene dosage effects! 3/
Comments
S100 family and MHC-II genes. #Echidna also disentangles intrinsic (CNA-driven) vs extrinsic expression immune signaling!
Read more here: https://www.biorxiv.org/content/10.1101/2024.12.15.628568v1
Let’s unravel tumor evolution, one clone at a time! 7/7