Cohesion directs homology search towards sister chromatid
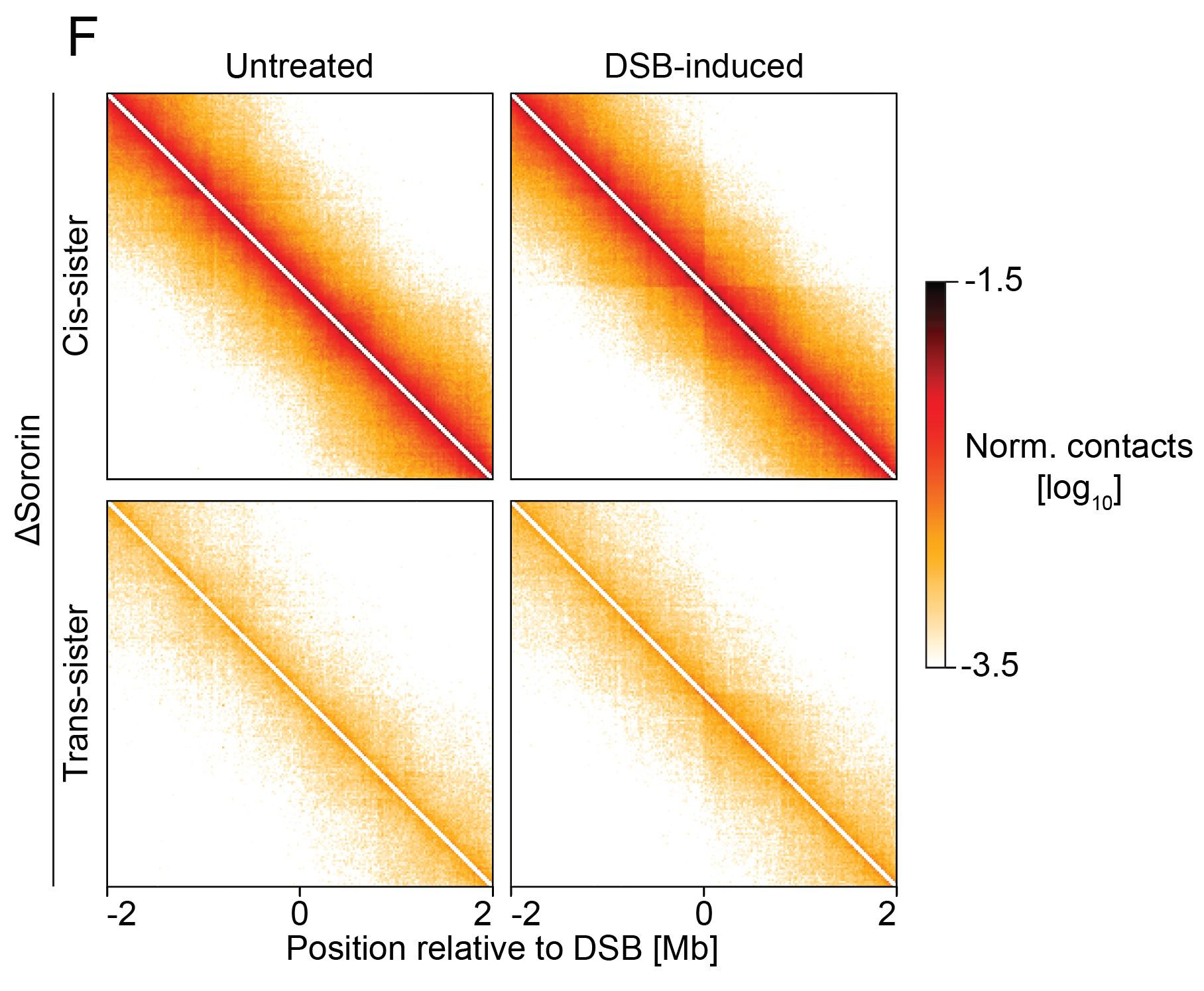
What happens if we disrupt sister cohesion? Upon sororin depletion (cohesion stabilizer):
✅ Loops remain intact
❌ Contacts between sister chromatids are lost
What happens if we disrupt sister cohesion? Upon sororin depletion (cohesion stabilizer):
✅ Loops remain intact
❌ Contacts between sister chromatids are lost
Comments
🔹Loop-forming cohesin defines search space, limited by TADs
🔹Cohesive cohesin tethers the break to its sister, to favour productive interactions
This ensures efficient & accurate homology search!
For more great science on this topic check the post below 👇
New method for monitoring homology search (RaPID-seq)? Preprint from @charlesyeh.bsky.social in @jcornlab.bsky.social! https://doi.org/10.1101/2025.02.10.63716
(BTW the DOI link is broken)
(Thanks for the DOI heads up, bluesky cut in the preview and it went unnoticed)
in that case, the sister chromatids are available without a genome-wide search only because synthesis has already taken place?