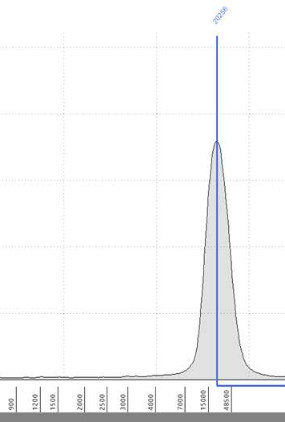
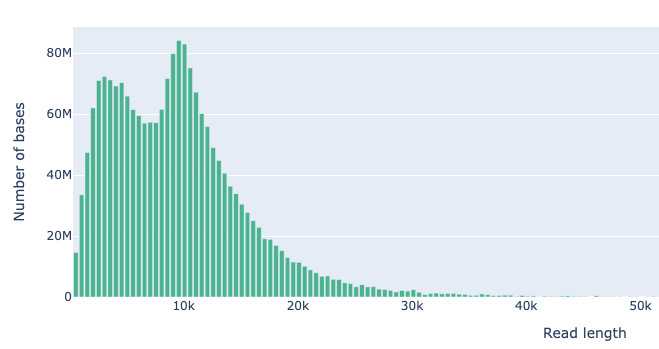
Oxford Nanopore metagenome read length weirdness - TapeStation of the input library shows very dominant peak at 20 kbp, what we actually got from the MinION was mostly 10kbp or lower (mean read length ~5kbp). This didn't happen for pure culture genomes on the same run. Any ideas why, anyone?
1 / 2
Comments
Meanwhile your nanopore data is a particle-by-particle DNA length distribution analysis.
Sage Science sells a power supply that lets you run pulse field in a std gel box; you can get good resolution for DNA up to at least 80kb (way beyond the reptation limit)
This is a very simple thing to do, although run times are bit long (~ 5+ hours)
https://sagescience.com/products/pippin-pulse/
There are some images at the end of the user manual
https://sagescience.com/wp-content/uploads/2021/06/Pippin-Pulse-User-Manual-460022-Rev-F-1.pdf