Stanford just found a natural alternative to Ozempic using some clever regex on the human proteome.
Instead of manually searching through proteins, their one-liner “peptide predictor” regex narrowed down promising candidates.
The calculation likely took just a few seconds.
Instead of manually searching through proteins, their one-liner “peptide predictor” regex narrowed down promising candidates.
The calculation likely took just a few seconds.
Comments
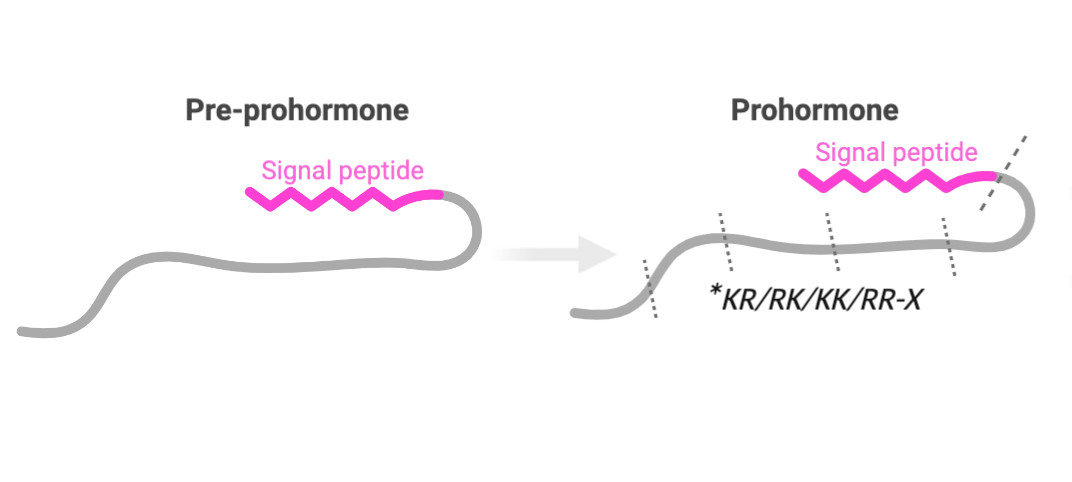
Here’s a breakdown of the regex one-liner:
- lookbehind to skip signal peptides
- scan for rare amino pairs (KK, KR, etc)
- prevent false cleavage match
- enforce minimum spacing + recursive pattern match
It’s a surgically accurate scalpel in the right hands. Tricky to master, but amazingly computationally efficient.
I wish more engineers would use it; the effort spent perfecting regex pays off dividends.