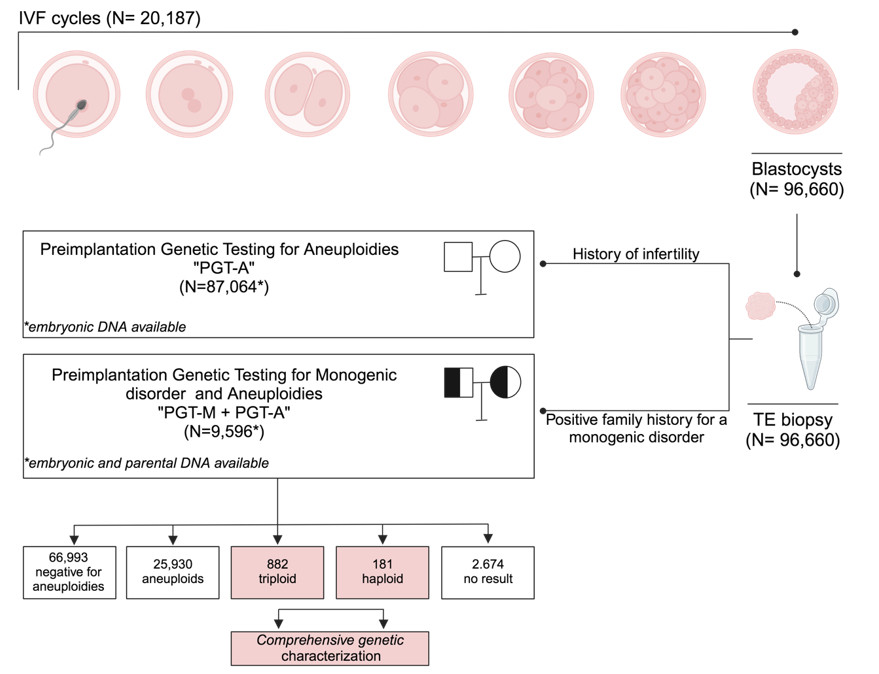
We started with 96,660 embryos that underwent PGT for aneuploidy using Juno's targeted sequencing method.
All were normally fertilized ("2PN") and developed to the blastocyst stage. We found 882 triploid embryos (an extra copy of all/most chrs) and 181 haploid (missing copy of all chrs).
3/9
All were normally fertilized ("2PN") and developed to the blastocyst stage. We found 882 triploid embryos (an extra copy of all/most chrs) and 181 haploid (missing copy of all chrs).
3/9
Comments
1) Sex chr composition (XXX/XXY/XYY etc)
2) 55 embryos with parental data
Findings:
1) Almost all triploid errors are maternal, with ~1/3 from meiosis I and ~2/3 from meiosis II
2) Almost all haploid errors are paternal
4/9
We found less crossovers than expected. Further, embryos with less crossovers had more additional aneuploidies in specific chrs (on top of the triploidy).
5/9
These embryos had both maternal chrs genome-wide.
(Except chrs with an additional aneuploidy; see figure for SNP array validation of one such embryo.)
6/9
Naturally, these embryos have high levels of homozygosity.
How the paternal genome disappeared is unknown.
7/9
Finally, ploidy aberrations tends to cluster in families: six couples had three affected embryos (four with female age<35), unexpected by chance, suggesting a genetic basis.
8/9
Identifying and understanding these errors will contribute to explaining meiosis and infertility.
9/9